Digital Discovery, 2023, 2,1452-1460
DOI: 10.1039/D3DD00088E, Paper
DOI: 10.1039/D3DD00088E, Paper
 Open Access
Open Access This article is licensed under a Creative Commons Attribution 3.0 Unported Licence.
This article is licensed under a Creative Commons Attribution 3.0 Unported Licence.David E. Graff, Edward O. Pyzer-Knapp, Kirk E. Jordan, Eugene I. Shakhnovich, Connor W. Coley
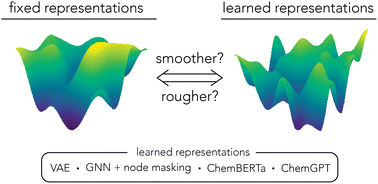
Pretrained molecular representations are often thought to provide smooth, navigable latent spaces; analysis by ROGI-XD suggests they are no smoother than fixed descriptor/fingerprint representations.
The content of this RSS Feed (c) The Royal Society of Chemistry
Pretrained molecular representations are often thought to provide smooth, navigable latent spaces; analysis by ROGI-XD suggests they are no smoother than fixed descriptor/fingerprint representations.
The content of this RSS Feed (c) The Royal Society of Chemistry